Protein-centric systems biology of the somatosensory neuraxis
Pain is complex, and our knowledge of the mechanistic underpinnings of pathological states (e.g., chronic pain) remains incomplete. Effective treatments are elusive, and centrally acting interventions still come with a plethora of side effects. A wealth of previous work has shown that the peripheral nervous system (PNS) is (i) a significant driver of chronic pain, and (ii) offers high therapeutic accessibility.
Enormous progress has already been made to implicate molecules, yielding important insights to genetic variants and relevant transcriptomic alterations in the PNS. To complement and extend these legacy data, we performed comprehensive protein profiling with data-independent acquisition mass spectrometry (DIA-MS) for three regions along the beginning of the mouse pain neuraxis: the sciatic nerve (SN), the dorsal root ganglia (DRG), and the spinal cord (SC). We compared these data to tissue proteomes from the mouse brain, enabling us to define "region-enriched" proteomes that is a set of proteins which are highly enriched in the SN and/or DRG (PNS-enriched) and in the SC, respectively (see: scheme of our workflow below). The following mouse regions were used for this comparison: the anterior cingulate cortex (ACC), the amygdala (AMY), the cerebellum (CER), and the prefrontal cortex (PFC).
The goal of our study was twofold. On one hand, we aimed to comprehensively define the adult mouse proteome of the SN, DRG, and SC. On the other hand, we envisioned using this information to enable the trustful monitoring of proteome alterations during a chronic pain condition. To address the latter, we examined protein changes in the PNS (DRG + SN) and spinal cord after the induction of neuropathic pain (spared nerve injury, SNI). For proteins detected in both pain and control conditions, results are presented as log2 fold changes, along with an adjusted p-value.
Methodologically, protein expression was examined for three replicates per condition (with 4 mice per replicate) and is presented as the average intensity across peptides and replicates. Peptides were only included in the analysis if they were detected across all replicates, and proteins were filtered for reviewed Uniprot IDs (see: http://www.uniprot.org/). Currently, all database entries are searchable by either Gene name (mouse) or reviewed Uniprot ID (mouse). Detailed methods, as well as tables including full analyses, region-enriched proteomes, protein changes after injury, and comparative analyses of our dataset with previously published data (mouse and human; genomic, transcriptomic, and proteomic) are available for download through our manuscript (Barry et al., 2018, Front. Mol. Neurosci., https://doi.org/10.3389/fnmol.2018.00259) and supplementary files.
User information:
- Individual query: all database entries are searchable by either gene name, protein name, or reviewed Uniprot ID (all mouse; see http://www.uniprot.org/). The dropdown menu in the search bar allows you to specify "search gene/protein name" vs "search (exact) ID" (Uniprot ID).
- If multiple entries appear, double click on the most interesting one for more information (ie. regulation upon SNI and tissue expression plots).
- Group queries: for genes with similar roots (ie. "Trp"), all corresponding entries will appear.
- Double click on individual entries for regulation upon SNI and tissue expression plots.
- Complete data query: enter a blank search.
- Region-enriched proteomes: Currently, there is no search function for this. We recommend checking out our manuscript (Barry et al., 2018, Front. Mol. Neurosci., https://doi.org/10.3389/fnmol.2018.00259): Full proteomes, region-enriched proteomes, and regulation upon SNI are supplied as supplementary tables.
- Significantly regulated proteins in SC, DRG, or SN: Choosing “show all signif SC/DRG/SN” and clicking the search button yields a table containing all significantly regulated proteins in the specified tissue.
Acknowledgements:
This work was supported by the Deutsche Forschungsgemeinschaft (Emmy Noether-Program: SCHM 2533/2-1 to MS; research grants: SCHM 2533/4-1 to MS, GO 2481/3-1 to DGV) and the Max Planck Society (PhD fellowship to JRS, student IMPRS fellowship to AMB).
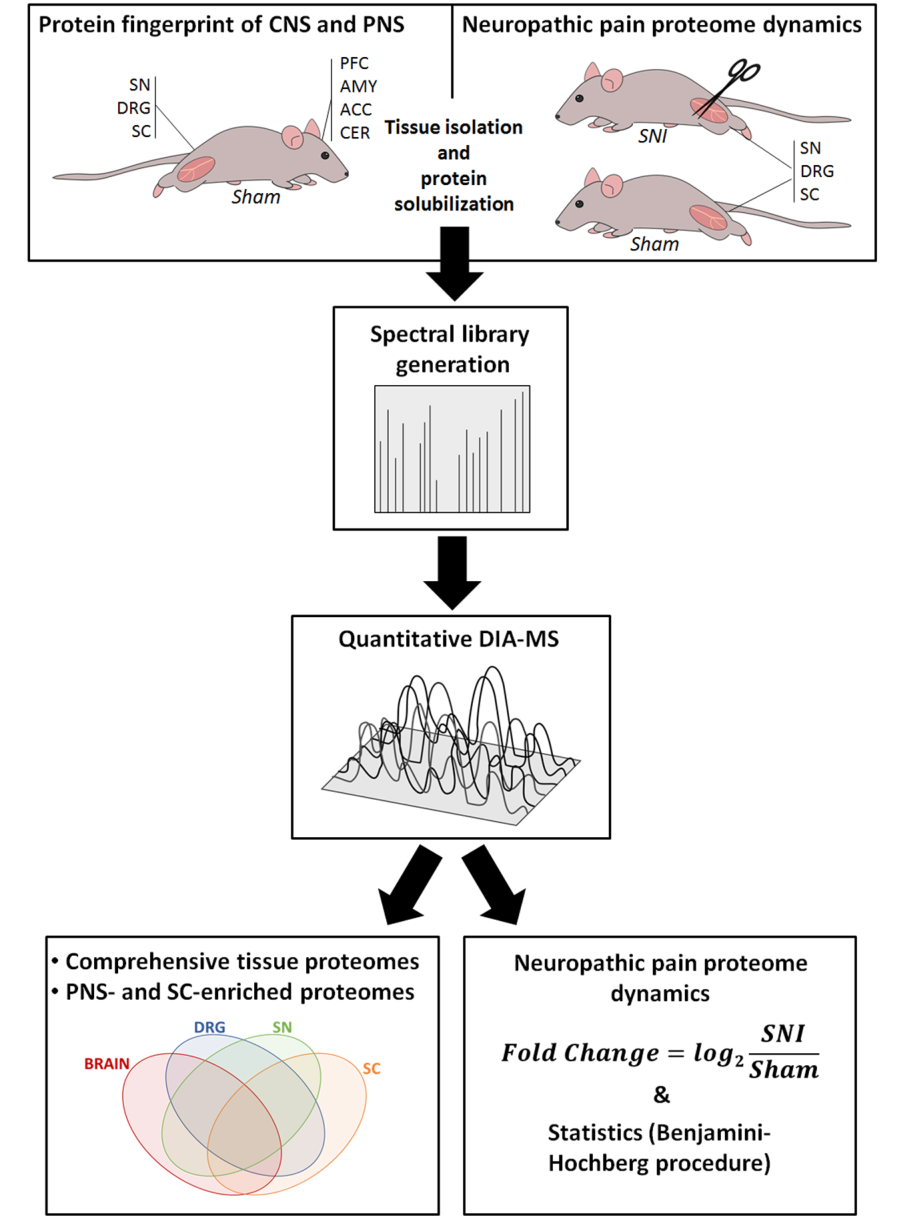
Figure legend: Overview of our integrated workflow: from the SNI model and Sham controls via mouse behavior, protein solubilization, quantitative DIA-MS and data analysis to region-enriched proteomes and proteome dynamics during pain. ACC, anterior cingulate cortex; AMY, amygdala; CER, cerebellum; DRG, dorsal root ganglia; PFC, prefrontal cortex; SC, spinal cord; SN, sciatic nerve.